The chromatin and histone proteins are responsible for compacting and organizing DNA within the nucleus and for regulating complex and dynamic patterns of gene expression.
Understanding the structure, mechanism and dynamics of the basic unit of chromatin – the nucleosomes – have gained increasing importance, aided by single molecule (SM) techniques. SM techniques enable to characterize molecular processes and identify the physical, chemical or biological sub-populations that are not accessible by ensemble studies.
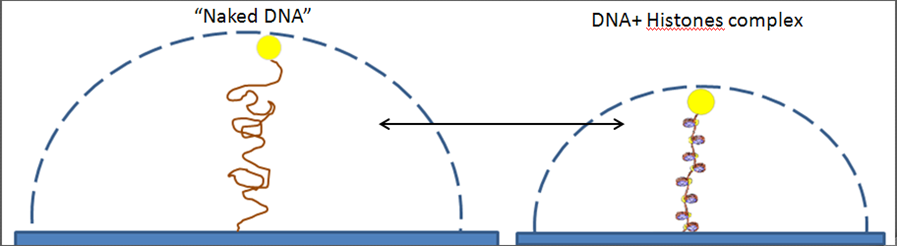
Illustration of nucleosomes studies using TPM.
The “naked” DNA (left) moves in a larger volume than the DNA with the histones (right).

TPM scatter plot of naked DNA (red) and DNA with histones (blue).
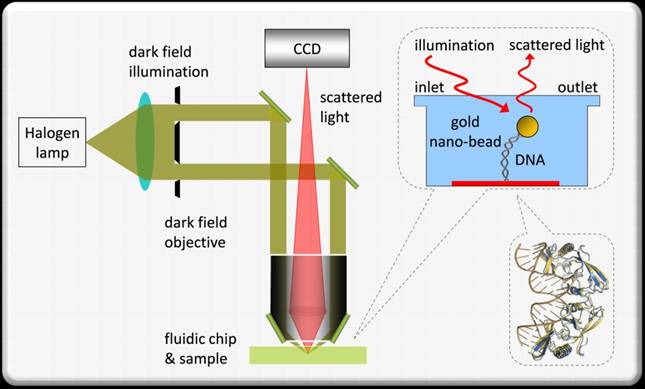
We use tethered particle motion (TPM) which is a rather simple and convenient SM technique. In TPM, a nano-bead is attached to one end of the DNA while the other end is attached to a glass surface. The DNA molecule is almost free to diffuse and to perform Brownian motion while we observe the scattering from the gold nano-bead. The distribution of the bead positions results from the physical properties of the polymer and a change in the distribution can be related to the bio-chemical reactions, such as nucleosome formation or annihilation.
Using TPM, we study the interaction of DNA and histone proteins, the dynamic process of nucleosome assembly/disassembly, the rate of these processes, and whether there is cooperation between the proteins.
Related paper:
- M. Lindner, G. Nir, H. R. C. Dietrich, I. T. Young, E. Tauber, I. Bronshtein, L. Altman and Y. Garini, Studies of single molecules at their natural form, Israel Journal of Chemistry 49, 283-291 (2009).