-
Using DNA Origami Structures for biophysical studies

DNA origami has been developed during the last decade for a large number of 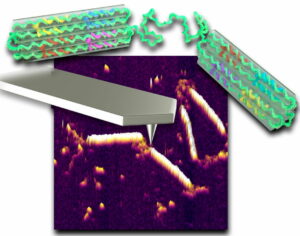 applications. We developed various DNA origami structures for studying the physical properties of DNA and for biosensing.
applications. We developed various DNA origami structures for studying the physical properties of DNA and for biosensing.
We study the structure and organization of the nucleus through the diffusion of chromatin loci. By combining Fluorescent microscopy measurements with the world of stochastic physics, we find the controlling mechanisms behind the motion of chromatin and the structure of the nanometer scale environment.
We study unfolding and refolding trajectories from single polypeptides chains, constructed of I27 domains from titin at the single molecule level without external forces using TPM. It allows us to determine single polypeptide chains biophysical properties during the unfolding and refolding.
 We are using Live-Cell imaging to study the genome dynamics in the nucleus, and to understand the telomeres’ properties of dynamics in live cells under different conditions (for example: disruption of telomeric proteins). Studying the nuclear dynamics enables us to understand the genome organization and various cell processes in vivo on a single molecule level.
We are using Live-Cell imaging to study the genome dynamics in the nucleus, and to understand the telomeres’ properties of dynamics in live cells under different conditions (for example: disruption of telomeric proteins). Studying the nuclear dynamics enables us to understand the genome organization and various cell processes in vivo on a single molecule level.
We believe that in the long-run, these studies will lead to further diagnostics and prognostic methods, as well as to the identification of drug-targets.
 Many intra-cellular processes are attributed to DNA-protein or RNA-protein interactions.
Many intra-cellular processes are attributed to DNA-protein or RNA-protein interactions.
To study such interactions, single molecule detection methods should be used. We aim to develop a simple system for 3D single molecule detection. The idea is to combine few methods, and get a simple 3D high-resolution system.
 We are studying this effect of HU using ‘minimal intervention’ methods that does not require the use of external forces, thus decreasing the interruption to the biological process.
We are studying this effect of HU using ‘minimal intervention’ methods that does not require the use of external forces, thus decreasing the interruption to the biological process.
We use Tethered Particle Motion (TPM) that involves the attachment of a single DNA molecule to a glass slide at one end, and the attachment of a gold nano bead to the other. The molecule diffuses in a restricted volume in the solution and the bead scattered light is detected with optical microscopy.
When a new nucleosome is created, it warps ~147 bp of the DNA, thus the end-to-end distribution of the molecule changes.
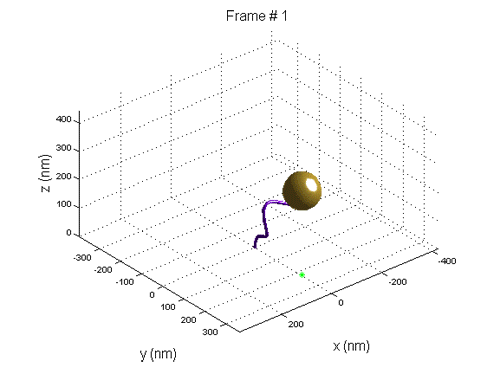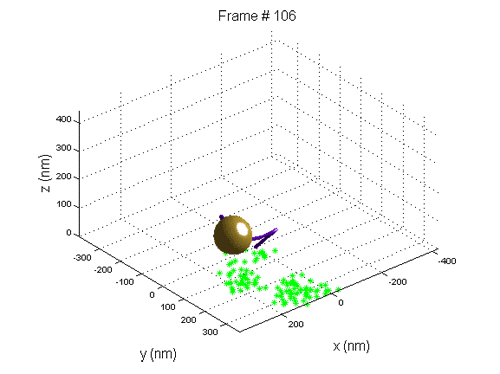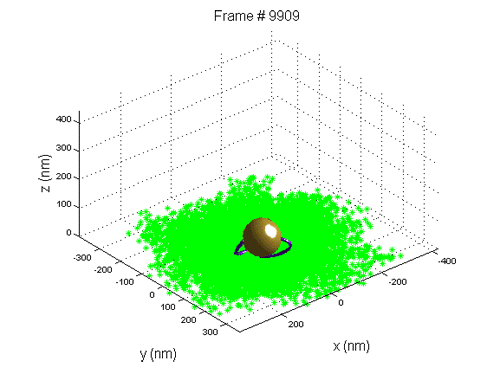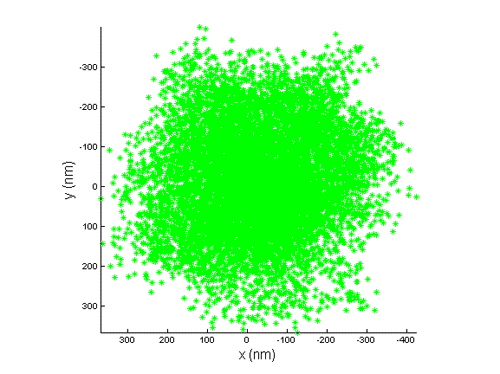
We use tethered particle motion (TPM) technique, that allows to measure the end-to-end distribution of a tested polymer such as DNA, for studying the binding interaction of DNA and histone proteins, the  dynamics of nucleosome assembly and disassembly with a resolution of one nucleosome, the rate of these processes, and whether there is cooperation between the proteins, thus the creation of one nucleosome promotes or inhibits the creation of another.
dynamics of nucleosome assembly and disassembly with a resolution of one nucleosome, the rate of these processes, and whether there is cooperation between the proteins, thus the creation of one nucleosome promotes or inhibits the creation of another.
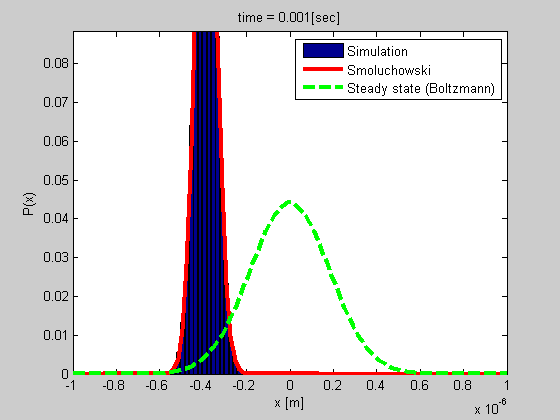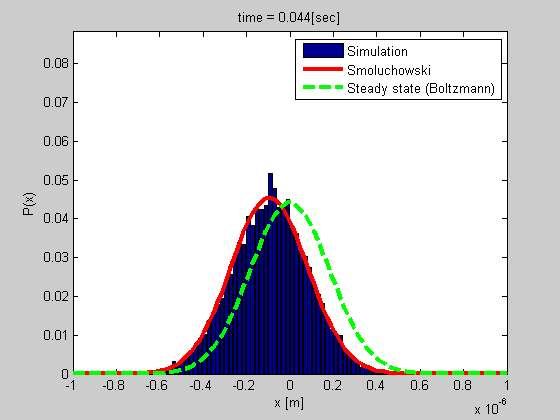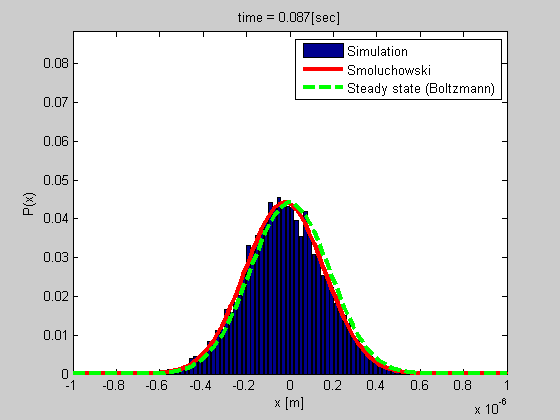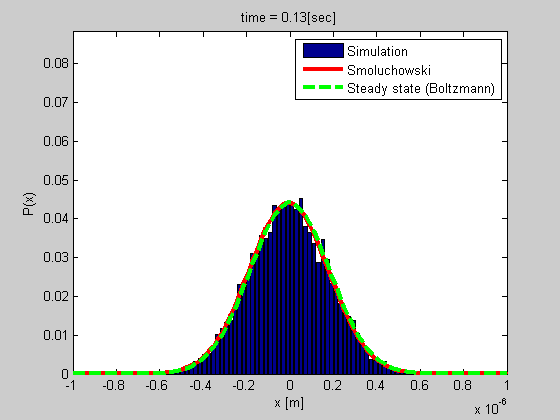 Many biophysical processes involves diffusing particles in a potential field, especially a harmonic potential. This is the case for a particle trapped in optical or magnetic tweezers, or a particle tethered to DNA.
Many biophysical processes involves diffusing particles in a potential field, especially a harmonic potential. This is the case for a particle trapped in optical or magnetic tweezers, or a particle tethered to DNA.
We developed a method for analyzing such a system using the Smoluchowski’s equation. The method is simple and do not require calibration, and it gives more data than most of the other current methods.
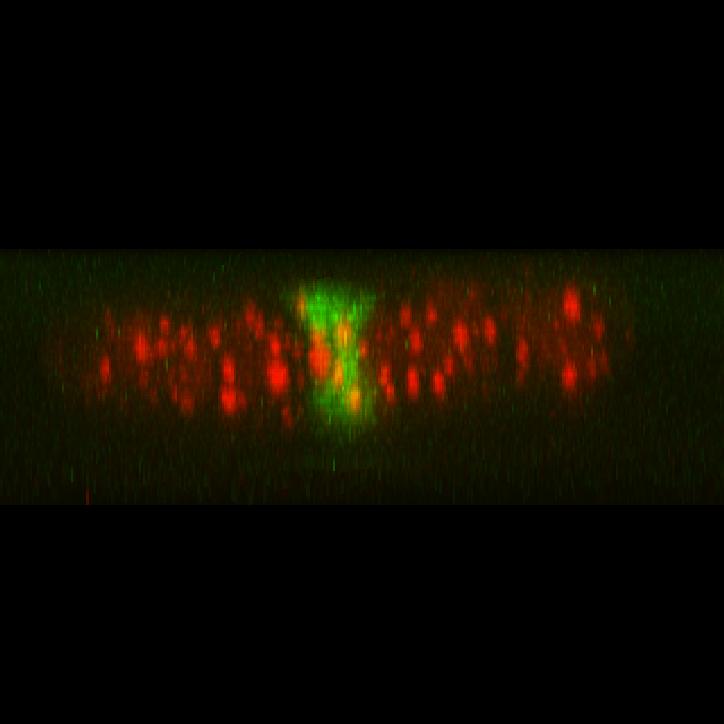
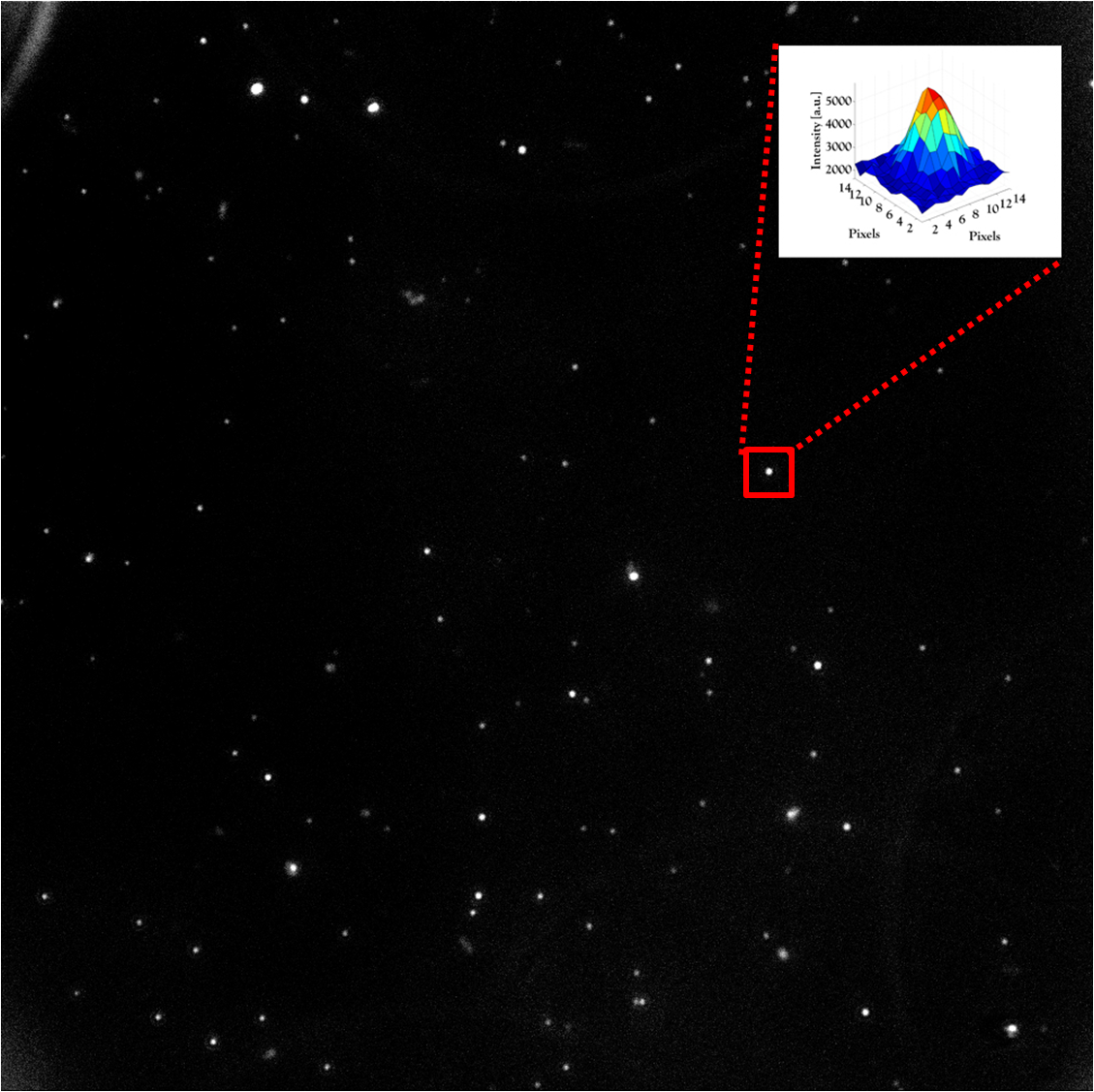
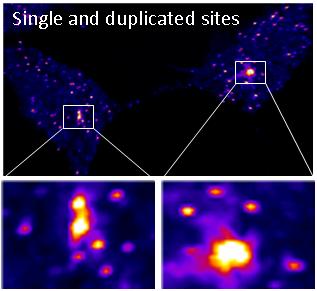
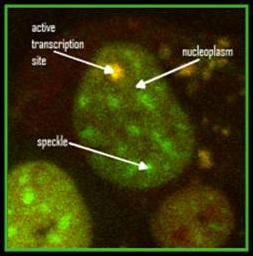
In collaboration with Dr. Yaron Shav-Tal’s lab, we developed a unique system to observe a single copy gene transcription integrated into the DNA of a human cell. With this system we can investigate transcription on a single-copy gene level in living cells.
Proteins can be viewed as nano-machines with moving elements. We are studying the link between conformational changes that a folded protein undergoes and its functionality at high temporal resolution.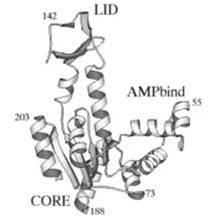
The model molecule presently used for this study is the E. Coli Adenylate kinase; a multi-domain enzyme molecule.
The research is performed at the single molecule level in high temporal resolution achieved by applying different fluorescence techniques such as FRET and FCS. The project is done as a collaboration with Haas’ group .
We are study protein dynamic using FCS (fluorescence correlation spectroscopy) method. With this technique we are study splicing factors fast dynamic (<1 sec) in living cells.